Km
The Michaelis constant, Km, is a parameter in the Michaelis-Menten equation. Km is equal to the substrate concentration where the corresponding reaction rate is ½ • Vmax. An enzyme with a low Km, therefore, achieves its half-maximal velocity at a low substrate concentration, while an enzyme with a high Km needs high substrate concentrations to achieve this velocity. It has been experimentally shown, that Km of an enzyme is usually close to the cellular concentration of its substrate. For an enzymatic reaction involving 2 steps, where the second step is rate-limiting, Km is approximately equal to the dissociation constant of the ES complex. In this case, a low Km implies a high affinity for the substrate; however, this interpretation of Km is only valid for a few enzymes [1].
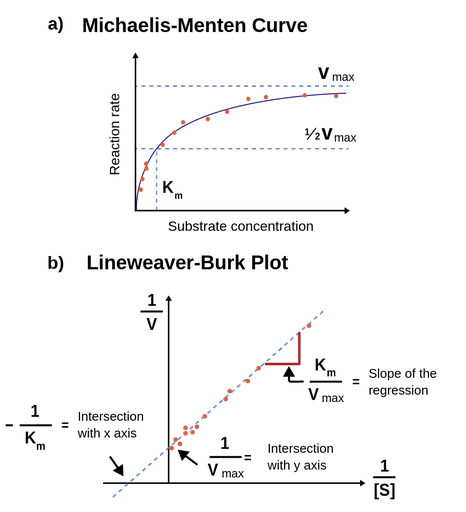
Figure 1: Figure 1a; Michaelis-Menten equation fitted to a plot of initial rates against substrate concentration. When the substrate concentration equals Km, the reaction rate is ½*Vmax. Figure 1b; An illustration of the Lineweaver-Burk equation fitted to a double-reciprocal transformation of an enzyme kinetic dataset.
The Lineweaver-Burk equation
Several methods for determining Km exist. The most direct method is to plot V0 against [
1/V0 = 1/Vmax + Km/Vmax • 1/[
This equation shows, that a plot of 1/V0 against 1/[
Other transformations
Notable, the Lineweaver-Burk transformation is not the only option. For example, a plot of [
References
-
Lehninger, Albert L.; Nelson, David L.; Cox, Michael M. (2008). Principles of Biochemistry (5th ed.). New York, NY: W.H. Freeman and Company. ISBN 978-0-7167-7108-1.
-
Atkins, G.L. and Nimmo, I.A. (1975) A comparison of seven methods for fitting the Michaelis-Menten equation. Biochem. J. 149, 775-777.